We’ve known for a long time that having kids with close relatives is bad—and a fascinating new paper puts a number to it. Being the child of cousins knocks more than three years off your life expectancy.
So what might that mean for longevity science?
Let’s start by taking a look at the results. Here’s a graph from the paper tweeted by one of the its authors, Munir Squires:
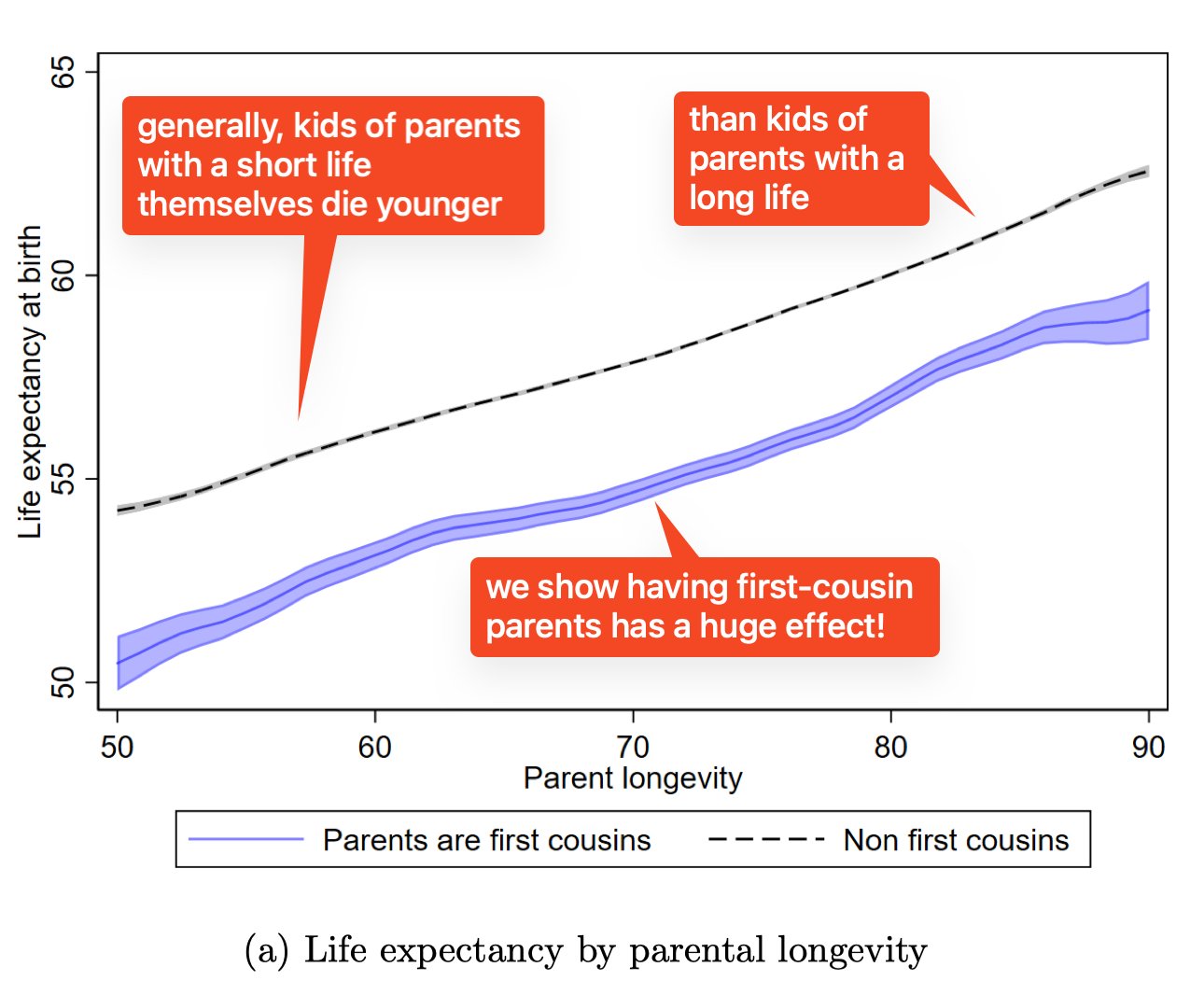
So children’s ages are correlated with their parents (albeit a bit weakly—every decade of parental longevity only adds a few years to theirs, because longevity isn’t purely genetic), and having parents who are first cousins knocks a few years off that life expectancy for all parental lifespans, which is an interesting result.
But this is an area of study fraught with potential confounding variables—might people who have kids with their cousins be systematically different in socioeconomic or other respects that mean their kids have shorter life expectancies? The authors try to control for this by only comparing children of siblings, one of whom married a cousin. Here’s a simple family tree explaining what they were comparing:
The idea is that brothers and sisters of people who have kids with their cousins will be very similar socioeconomically, genetically and more, removing a lot of the potential confounding factors. And, according to their tests, this seems to work! Since this obviously isn’t something we can do a randomised trial of, this level of evidence might be the best we can do.
I bet there’s something interesting in the genetics here for longevity research to dig into. Lifespan is probably dictated by hundreds or thousands of genes, and the problem with ‘consanguinity’ (which is the technical and slightly more polite term for inbreeding) is that ‘recessive’ genes are more likely to be present in two copies, one from each parent. Recessive genes are those where a single copy is unproblematic (so the parents might not have any symptoms) but, in the extreme case, the kid with two copies might have a genetic disease.
While specific recessive diseases are the most obvious issue, this life expectancy difference could also be driven by far more subtle effects from dozens of recessive genes making everything run slightly less well throughout life. It would be interesting to dig into the specific causes of death and DNA sequences in a population like this, and see if we can pick up any patterns that are related to the ageing process more generally.
This is also interesting context for a study that discovered a gene in the Old Order Amish that allowed people with one copy to live ten years longer than those without. The gene was only discovered because an Amish girl inherited two copies, one from each parent, and it caused her blood not to clot. After many years of detectivework, it was found that people with only one copy (like her parents!) lived a decade longer.
Here’s an extract from my book, Ageless, about this discovery:
This is the chain of events that led a three-year-old girl, part of the Old Order Amish community in Berne, Indiana, to hospital in the mid-1980s—a trip which would eventually lead to the discovery of a new gene for human longevity. After bumping her head, the girl developed a large pool of blood under her scalp, and surgery to drain it only made things worse—she nearly bled to death. A few years later, surgery for a dental abscess nearly caused her to drown in her own blood. There’s a collection of ‘bleeding disorders’ which could be responsible for blood not clotting to seal up a wound but, one by one, doctors eliminated them. This girl’s condition was something that, at that time, was unknown to medical science. It was only due to the persistence of a doctor and blood-clotting specialist named Amy Shapiro that the underlying cause of her bleeding was uncovered.
Trawling the literature for clues, Shapiro read about a protein called PAI-1 which is involved in blood clotting. She eventually managed to persuade a colleague to test the sequence of the gene that codes for it, SERPINE1, in the girl’s DNA. They found a two-letter mistake.
…
In 2015, almost two hundred Amish people volunteered for a battery of medical tests examining their blood and heart health. Those with a single copy of the mutated SERPINE1 gene had slightly better cardiovascular health than those with two copies of the regular gene along, interestingly, with longer telomeres. They were also much less likely to get diabetes—eight of the 127 non-carriers had diabetes, against zero of the 43 with the mutation. Most strikingly, using genetic testing and family trees to deduce the genotype of relatives who had already died, the study found that people with the mutation lived ten years longer on average than those with two copies of the normal gene, boosting average lifespan from 75 to 85 years.
Given that her parents, and dozens of other Amish people with a single copy of the gene, had no overt symptoms, consanguinity was what allowed this gene to be discovered…but might also mean that Amish life expectancy is reduced in general because of this, and/or this gene might be in some way compensating for its effects, making the ‘10-year life extension!’ less impressive.
The good news is that life expectancy of Amish without the mutation is 75, and with one copy that’s boosted to 85—if it was boosting it from 60 to 70 or something, that would be less exciting. And, to oversimplify, 10 years is more than the 3-year consanguinity penalty so that’s good too!
We should still be cautious because the SERPINE1 gene the found might be doing something that’s specific to the genetics or lifestyle of the Amish, so maybe I’ll hold off on having my DNA edited for now! But I’m still cautiously optimistic about it as a human longevity gene.
And the study we started out discussing provides another way that the genetics of isolated communities might give us a window into the genetics of human longevity.